Accessing the PolyHub Data Archive
All data is saved under the $DATA grid variable for the standard PolyHub grid site. At the main PolyHub site (The Newton cluster at The University of Tennessee), this directory is /data/EVO/ . Organization of the data archive is based on simulation level, polymer structure, number of carbon and so on. Figure 1 displays the structure of our data archive. For example, data sets containing the properties of linear C50H102 liquids at shear rate 1.0 are located at
$DATA/Atomistic -> Linear -> c50 -> shear -> c50_Shear_1.0
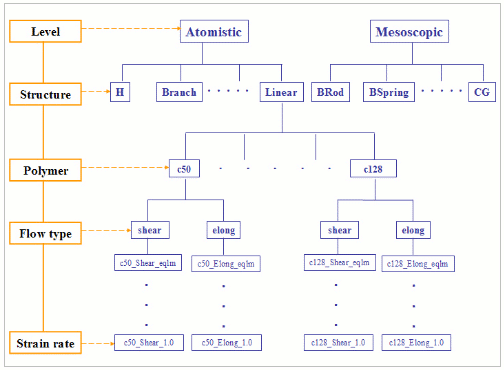
Figure 1: PolyHub Data Archive Structure
Models and Schemes
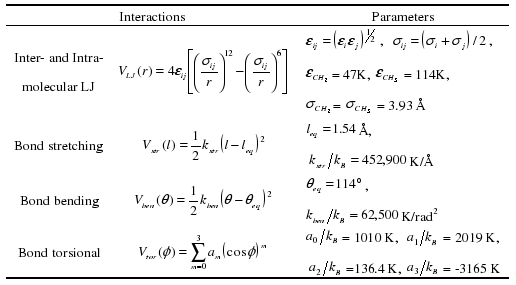
The non-equilibrium molecular dynamics (NEMD) simulation of linear alkanes was performed in the NVT ensemble using p-SLLOD equations of motion for a homogeneous flow. The Nosť-Hoover thermostat is added to control temperature of the system. For shear flow, the Lees-Edwards boundary condition is applied while the Kraynik-Reinelt boundary condition is used for elongation flow. The Siepmann-Karaboni-Smit (SKS) united-atom model for linear alkanes was adopted because of extensive usage for simulating rheological behavior of alkanes and polyethylenes. But, for bond-stretching interaction, the rigid bond between adjacent atoms was changed to a harmonic potential function. The four types of interactions are summarized in Table 1. The equations of motion were integrated using r-RESPA. The simulations were carried out at the temperature of 450K. In order to prevent the system-size effect, the box dimensions is carefully chosen to consider the fully-stretched length of linear alkanes in both flows.
Data archive contents
- Input data
input.dat: parameters like run type, simulation length, temperature, shear rate etc.
pos.dat: initial configuration of molecules, the total number of particle and chain, box size etc.
mdconfout.dat: last configurations of molecule - Output data
chainform.dat: mean-square end-to-end distance, mean-square radius of gyration
ct.dat: conformation tensor (Cxx, Cxy, Cxz, Cyy, Cyz, Czz)
uct.dat: unit conformation tensor (Uxx, Uxy, Uxz, Uyy, Uyz, Uzz)
energy.dat: total, potential, and kinetic energy
pt.dat: pressure tensor (Pxx, Pxy, Pxz, Pyy, Pyz, Pzz)
previs.dat: total, potential, and kinetic pressure, total, potential, and kinetic viscosity